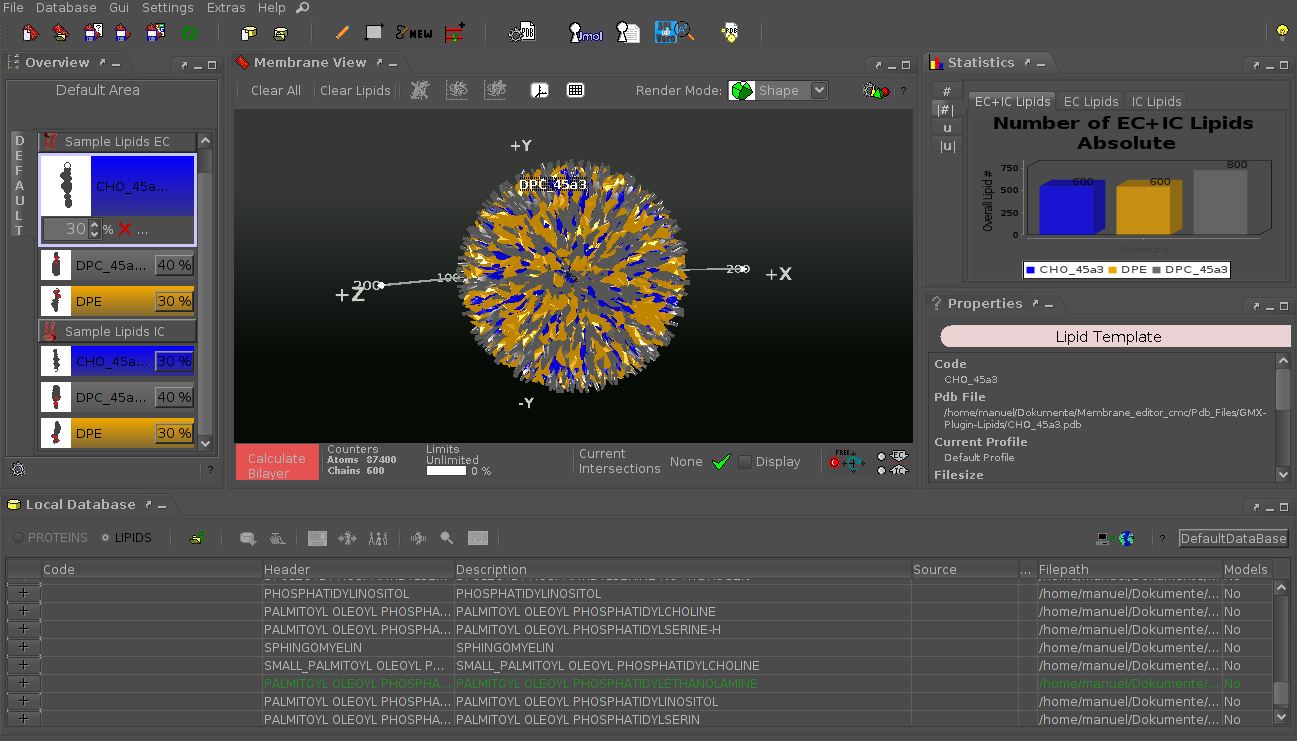
The CELLmicrocosmos 2 MembraneEditor is a 3D software which can be used to model molecular membranes based on the PDB format. These model membranes can be used to visualize biomolecular processes or to create base membranes as starting point for molecular dynamic simulations. Those simulations can be used to simulate membrane-protein or drug-membrane interactions. Rectangular membranes, membrane segments, single- or double-layer membranes, and also vesicles can be created by using this software.
It’s a Java-based software which can be run with professional 3D-stereoscopic displays as well:
http://Cm2.CELLmicrocosmos.org
The latest paper published was covering the Vesicle Builder plugin for the MembraneEditor:
The Vesicle Builder-A Membrane Packing Algorithm for the CELLmicrocosmos MembraneEditor
B Giuliari, M Kösters, J Zhou, T Dingersen, A Heissmann, R Rotzoll, J Krüger, A Giorgetti, B Sommer
The Eurographics Association: Workshop on Molecular Graphics and Visual Analysis of Molecular Data 2020
[Conference Article]
2020
Also part of this project, a new version of APL@Voro was published in 2023. This tool can be used to analyze membranes created by the MembraneEditor and then simulated with Gromacs – a Molecular Dynamics Simulation tool:
APL@voro—interactive visualization and analysis of cell membrane simulations
M Kern, S Jaeger-Honz, F Schreiber, B Sommer
Bioinformatics 39 (2), btad083
[Journal Article]
2023
Project Leader: Dr Bjorn Sommer
Collaboration Partners: Prof Dr Falk Schreiber